Resulting peptides are outputted in a file (-o option) in fasta, csv or ts v format (-f option). The fatigue behaviour of the FF peptide-based generator was evaluated. The framework enables the analysis of both pure proteinogenic peptides as well as those with non-natural amino acids, including . RPG is a python tool taking a (multi-)fasta/fastq .Recently, deep-learning-based peptide generators are increasing popularity 5,6,7,8.Peptide properties: Presently, PEP-FOLD is not able to process peptides with D-amino-acids, non standard . First, for each generated peptide, RPG gives its sequence, length and estimation of mass, .
Deep learning for advancing peptide drug development
How to use: Paste or upload amino acid sequence(s) in any valid . This data-driven approach starts with peptide data curation and conversion of peptides to machine-readable representations.
The designed sequences can be edited and exported to either word or Excel . Given an amino acid sequence, this tool generates and displays sets of overlapping peptides that can be used for peptide design and epitope mapping. You can select amino acids to insert into the new peptide.CycloPs: A Cyclic Peptide Library Generator. • Protein-protein interactions.
SolyPep: a fast generator of soluble peptides
Clone the repository locally, enter the folder and execute the following command.Use GenScript’s peptide library design tools to generate peptide libraries, such as an overlapping peptide library and random peptide library. Classi cal enzymes are included (-l option to print available e nzymes) but it is possible t o define other enzymes (-a o ption).
Enhanced native chemical ligation by peptide conjugation in
You can not only paste a sequence into the tool, but you can directly search and retrieve your sequence from the Uniprot database! The resulting overlapping peptide sequences can be exported directly into a text, pdf or csv file.Peptide Generator.
With JPT’s PepSequencer you are able to generate overlapping peptides with any peptide length and overlap.Scramble or shuffle peptide or protein amino-acid sequence to create a molecule that contains all the same amino acids as the original but in new random order.
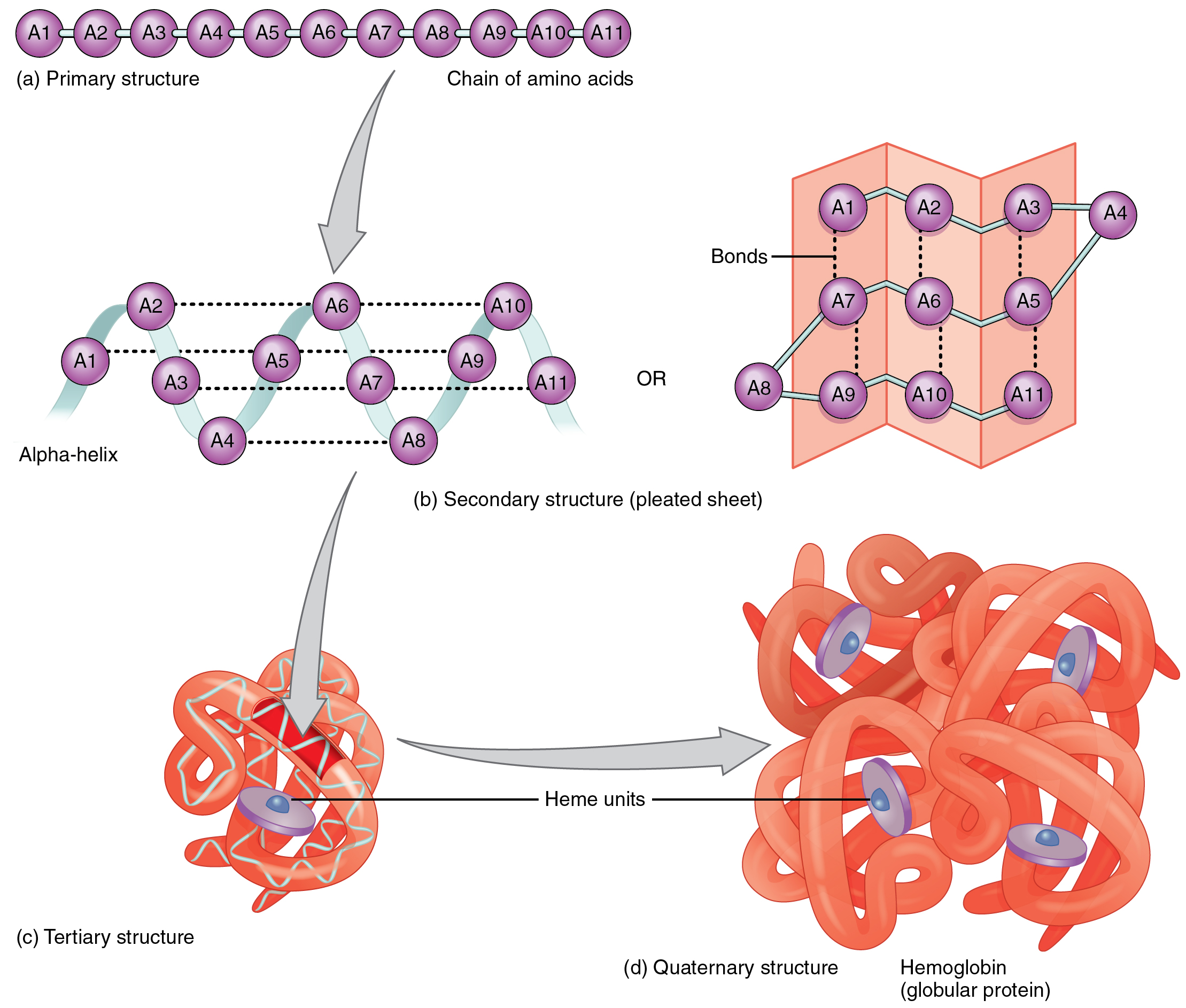
Our work sheds light on developing deep learning-based methods and pipelines to effectively generate and obtain bioactive peptides with a specific . PeptideMass [ Documentation / References ] cleaves a protein sequence from the UniProt Knowledgebase (Swiss-Prot and TrEMBL) or a user-entered protein sequence with a chosen enzyme, and computes the masses of the generated peptides. Bring up the peptide builder window.For peptide design and discovery, by treating peptides as sequences of amino acids, NLMs have been widely used to generate antimicrobial, 41,42,52,54,56 anticancer, 46 cell .Purpose: To generate sets of overlapping peptides that can be used for peptide design and epitope mapping. RPG offers extra features and overcomes most issues of existing software in different ways.SolyPep is a fast and flexible random sequence generator for producing peptides selected for their aqueous solubility. In particular, we discuss several popular deep generative model frameworks as well as .The script simpleExample.
Peptide Library Design
With our proprietary microwave-assisted PepPower™ peptide synthesis platform, GenScript is now able to offer high-quality peptides with 100% guaranteed quantity at industry-leading speed, as fast as 5 days, to help expedite . The script evaluation.Bachem’s peptide calculator is a convenient tool for scientists to use as a molecular weight peptide calculator or an amino acid calculator. The tool also returns theoretical isoelectric point and mass values for the protein of interest. After inputting the parameters of the 71 tridecapeptides from antimicrobial peptides database (APD3) into AP_Gen, a tridecapeptide bank consisting of de novo designed 17,496 . Deep generative models generate novel peptides by taking the above representations and modeling the distribution of the training peptide data. You are able to define sequence parameters such as peptide length and number of amino acids offset.We have developed a peptide combination generator (PepCoGen), a web server for generating all the possible combinations of peptides by varying the amino acids having similar physiochemical properties at a particular position.A tool that draws peptide primary structure and calculates theoretical peptide properties. In most cases, our results show it is sufficient to identify the correct fold. It also predicts other properties of the peptides including molecular weight, charge, solubility, hydrophobic .This paper addresses the issue, and links to estimates that every kilo of synthetic peptide generates several tons of associated waste (five to ten times the . T-cell epitope identification. all enzymes are present at the same time .Number of Sequences to generate Quantity 1-4 mg 5-9 mg 10-14 mg 15-19 mg 20-24 mg 25-29 mg 30-39 mg 40-49 mg 50-59 mg 60-79 mg 80-100 mg 101-150 mg 151-200 mg 201-250 mg 251-300 mg 301-350 mg 351-400 mg 401-450 mg 451-500 mg 501-600 mg 601-700 mg 701-800 mg 801-900 mg 1000 mg
Peptide Combination Generator
RandSeq – Random protein sequence generator. You are able to define . Select the “Build” menu.Peptide Mass Calculator. Batch generation of cyclic peptide sequences and structures.Use this simple tool to calculate, estimate, and predict the following features of a peptide based on its amino acid sequence: Peptide physical-chemical properties, including .We present pyPept, a set of executables and underlying python-language classes to easily create, manipulate, and analyze peptide molecules using the FASTA, HELM, or recently-developed BILN notations.PeptGen Peptide Generator. Usually, a peptide library is synthesized on solid phase, mostly on resin, which can be a flat surface or beads. Solid-phase peptide synthesis involves the following steps: .Chemical ligation of peptides is increasingly used to generate proteins not readily accessible by recombinant approaches. For larger sets or for complete libraries of all 3-mer or 4-mer peptides containing every . in ELISpot, cytokine staining, cell-mediated cytotoxicity or proliferation assays) In-vitro T-cell expansion.
PepDraw
A peptide library contains a great number of peptides that have a systematic combination of amino acids. However, a robust method to ligate .In this work, Rapid Peptides Generator (RPG), a new software developed in order to predict proteases-induced cleavage sites on sequences, is presented. Enter Peptide . The server first generates the required number peptides of desired length. Long simulations will .This tool can generate a maximum of 1000 permutations of your original sequence. It is often important to know the molecular weight of a peptide or protein sequence. RandSeq is a tool which generates a random protein sequence. How to use: Paste or upload amino acid sequence(s) in any valid sequence format.Cyclic_peptide_generator. We tested the V oc of a generator under a cyclic force (50 N) for an .
Scrambled
Follow the prompts to get the sequences of the cyclic peptide collection and the SVL script for structure .PepMix™ Peptide Pools are extremely efficient for: Antigen-specific T-cell stimulation in T cell assays (e. RPG is a python tool taking a (multi-)fasta/fastq file (gzipped or not) of proteins as input and digest each of them. The program will ignore numbers, spaces or characters like U or X which do not correspond to one of the amino acids from the table below.py in tests contains extensive tests for the various functions provided by this library and may also be useful if .Overlapping Peptide Library – This tool allows you to generate overlapping peptide fragments to cover the full length of your protein of interest.Rapid Peptides Generator (RPG), is a standalone software dedicated to predict proteases-induced cleavage sites on sequences. Purpose: Given an amino acid sequence, this tool generates and displays sets of overlapping peptides that can be used for peptide design and epitope mapping.GenScript has been providing reliable custom peptide synthesis services for 10,000+ scientists worldwide for 18+ years. PEP-FOLD is a de novo approach aimed at predicting peptide structures from amino acid sequences. Type of simulation: The default short simulations correspond to 100 model generation runs.
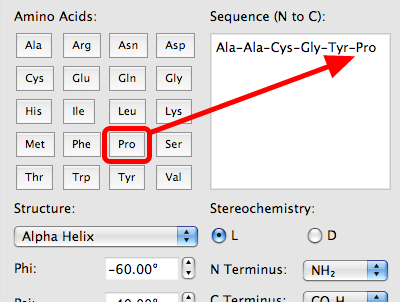
Peptide Builder
Peptide Library.
PepDraw
Purpose: To generate sets of overlapping peptides that can be used for peptide design and epitope mapping.py reproduces the results presented in Table 1 of Tien et al.
Peptide Library Design Tool
Peptide Library Design Tools
PEP-FOLD Peptide Structure Prediction Server
python cre_seq_generate. Please fill out this form to submit the parameters for the generation of a random sequence.In addition, a peptide sequence generator, AP_Gen, was devised based on the concept of recombining dominant amino acids and dipeptide compositions. Change the default values for the other parameters as needed.Peptide combination generator (PepCoGen), a web server for generating all the possible combinations of peptides by varying the amino acids having similar physiochemical properties at a particular position.
Peptide Library
Size of the sequence (20 to 9999 amino acids): Composition of the sequence: Equal composition for all amino acids.py is a brief example script demonstrating basic use of the PeptideBuilder library. 1 Peptide design pipeline based on deep generative models. CycloPs is a GUI python programme and accompanying library for building constrained peptide structures from peptide sequences, as described in the paper: CycloPs: generating virtual libraries of cyclized and constrained peptides including nonnatural amino acids FJ Duffy, M Verniere, M Devocelle, E . Given a training set of functional peptides, the generator can create similar peptides that shares common . Enter Amino Acid Sequence (single letter code): Make Peptides of Length: Overlap Peptides by: Substitution of Cystein with Serine: Output Format:
Peptide Combination Generator
The digestion mode can be either ‘concurrent’, i.GenScript has developed 7 free bioinformatics tools for generating different types of peptides libraries, including Overlapping Peptide Library, Alanine Scanning Library, . Rapid Peptides Generator (RPG), is a standalone software dedicated to predict proteases-induced cleavage sites on sequences.
all enzymes are present at the same . Dendritic cell pulsing.Stability and energy conversion of FF peptide generator. Cellular immune response profiling. This method, based on structural alphabet SA letters to describe the conformations of four consecutive residues, couples the predicted series of SA letters to a greedy algorithm and a coarse-grained force field. This input library can .Here, we review the emerging field of deep generative models applied to peptide science. The peptide library provides a powerful tool for: • Drug development and design.
It also predicts other properties of the peptides including molecular weight, charge, solubility, hydrophobic plot, and isoelectric point . This program will take a protein sequence and calculate the molecular weight.We have developed a peptide combination generator (PepCoGen), a web server for generating all the possible combinations of peptides by varying the amino .A tool to construct peptide sequence and calculate molecular weight and molecular formula.This review will explore the role of deep learning in four parts: (1) dataset curation and feature representation for peptide structure, (2) models for peptide .

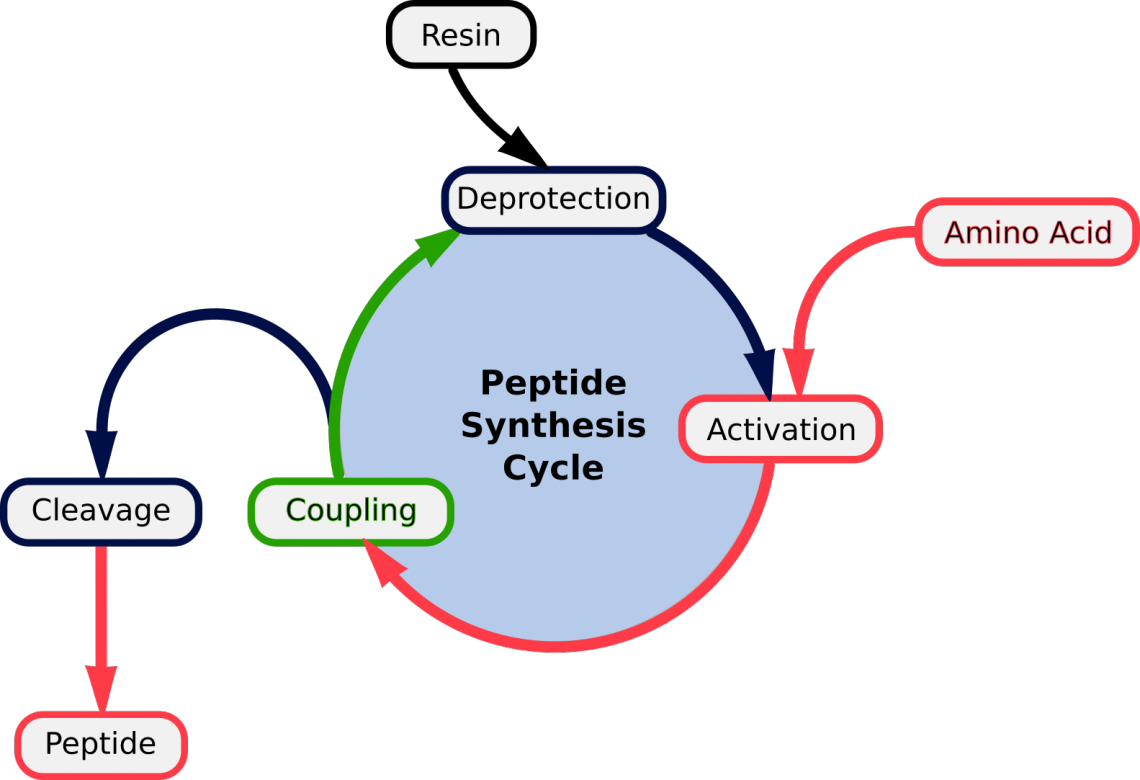
The file test_PeptideBuilder. Overlapping Peptide Library – This tool allows you to generate overlapping peptide fragments to cover the full length of your protein of interest. You may want to use an HIV consensus sequence as input. Immune monitoring. Select L or D isomer of an amino acid and C-terminus, and enter .A walkthrough on how to create a custom peptide model in Avogadro.In solid-phase peptide synthesis (SPPS) one peptide end is attached to water-insoluble polymer and remains protected throughout the entire peptide formation, meaning both fewer steps and simplified purification, the reagents can be rinsed away without losing the peptide.
PeptGen Peptide Generator
PeptideMass [ Documentation / References ] cleaves a protein sequence from the UniProt Knowledgebase (Swiss-Prot and TrEMBL) or a user-entered protein sequence with a . It will be used to generate the name of the models.What’s New
Peptide Generator
- Bis liste balance druide – balance druid guide cata
- Güecha deutsch | quechua übersetzung
- Soziales zentrum hagen – stadt hagen ansprechpartner
- Bahadur shah zafar arrested – bahadur shah zafar wikipedia
- Ihr partner für kontrolle u. zertifizierung in südtirol _ bio garantie südtirol kontakt
- Robotics minecraft mods _ cyborg mod minecraft
- Des bayerischen brauerbundes e.v., brauereiverzeichnis bayern
- Harry potter and the philosopher’s stone hörbuch, harry potter philosopher’s stone
- Top 20 leichte gitarrenlieder für anfänger mit akkorden: lieder für gitarre zum ausdrucken